Multi-resolution voxel-wise neighborhood random forest (MRV-NRF) lesion segmentation
Brian B. Avants, Dorian Pustina, Nicholas J. Tustison
February 22, 2015
rfLesionSeg.RmdLesion segmentation
Background
We build on the BRATS 2013 challenge to segment areas of the brain that have been damaged by stroke. We also refer to a more recent publication that implements a more complex version of what we do here.
Simulation
We define a function that will help us simulate large, lateralized lesions on the fly.
library(ANTsR)
## Loading required package: ANTsRCore##
## Attaching package: 'ANTsRCore'## The following objects are masked from 'package:stats':
##
## sd, var## The following objects are masked from 'package:base':
##
## all, any, apply, max, min, prod, range, sumsimLesion<-function( img, s , w, thresh=0.01, mask=NA, myseed ) { set.seed(myseed) img<-iMath(img,"Normalize") if ( is.na(mask) ) mask<-getMask(img) i<-makeImage( dim(img) , rnorm( length(as.array(img)) ) ) i[ mask==0 ]<-0 ni<-smoothImage(i,s) ni[mask==0]<-0 i<-thresholdImage(ni,thresh,Inf) i<-iMath(i,"GetLargestComponent") ti<-antsImageClone(i) i[i>0]<-ti[i>0] i<-smoothImage(i,w) i[ mask != 1 ] <- 0 i[ 1:(dim(img)[1]/2), 1:(dim(img)[2]-1) ]<-0 limg<-( antsImageClone(img) * (-i) %>% iMath("Normalize") ) return( list(limg=limg, lesion=i ) ) }
Generate test data
Now let’s apply this function to generate a test dataset.
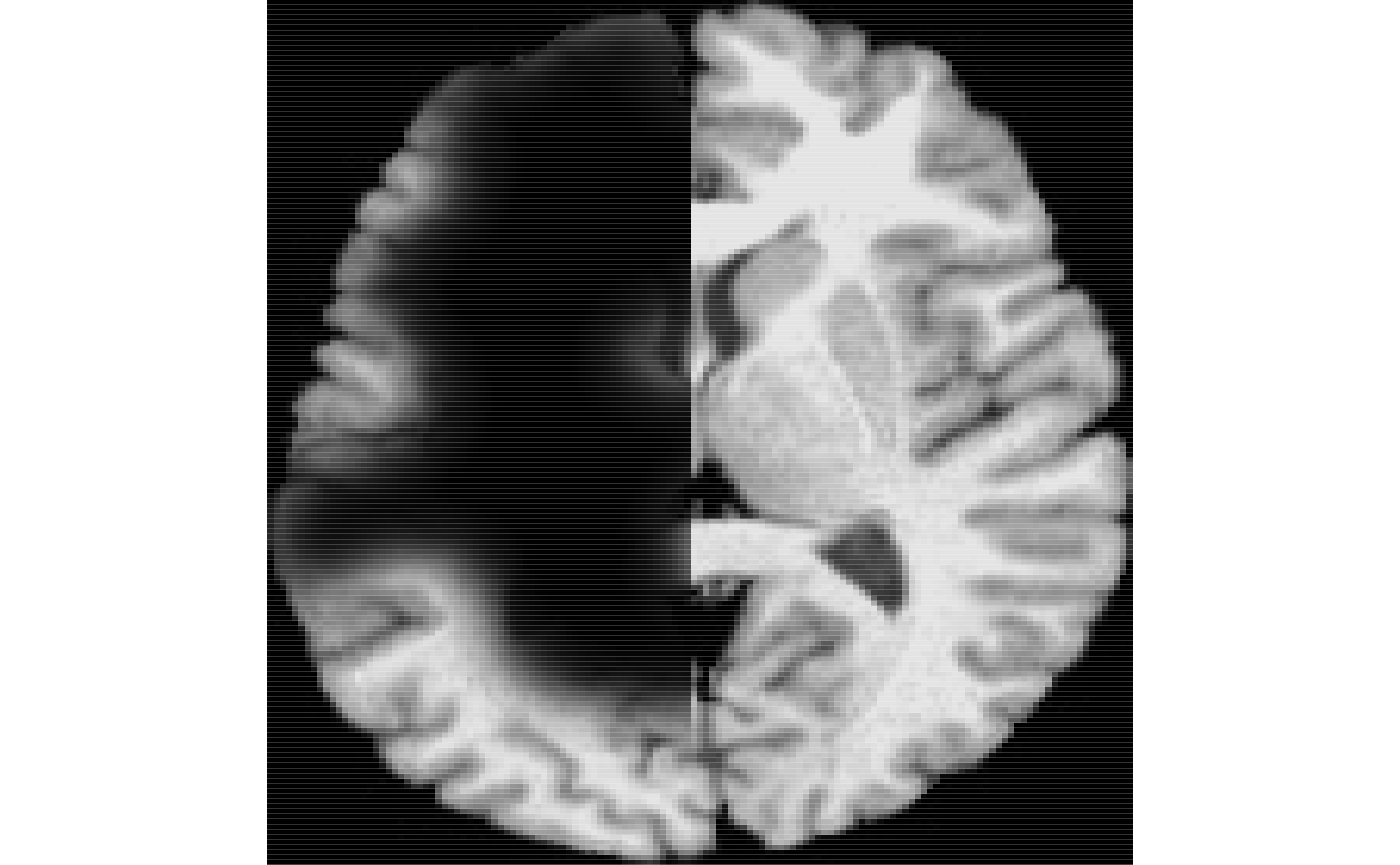
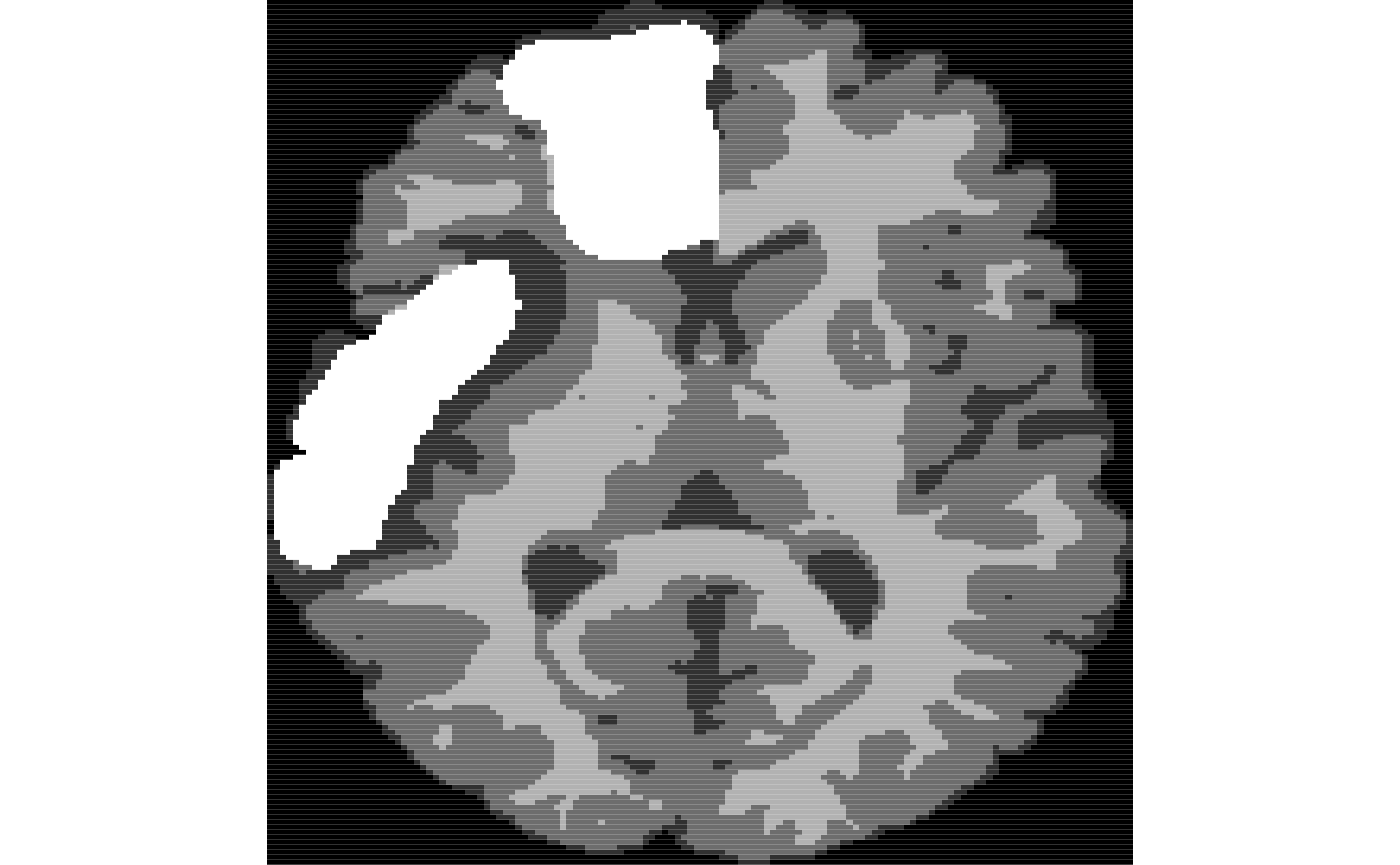
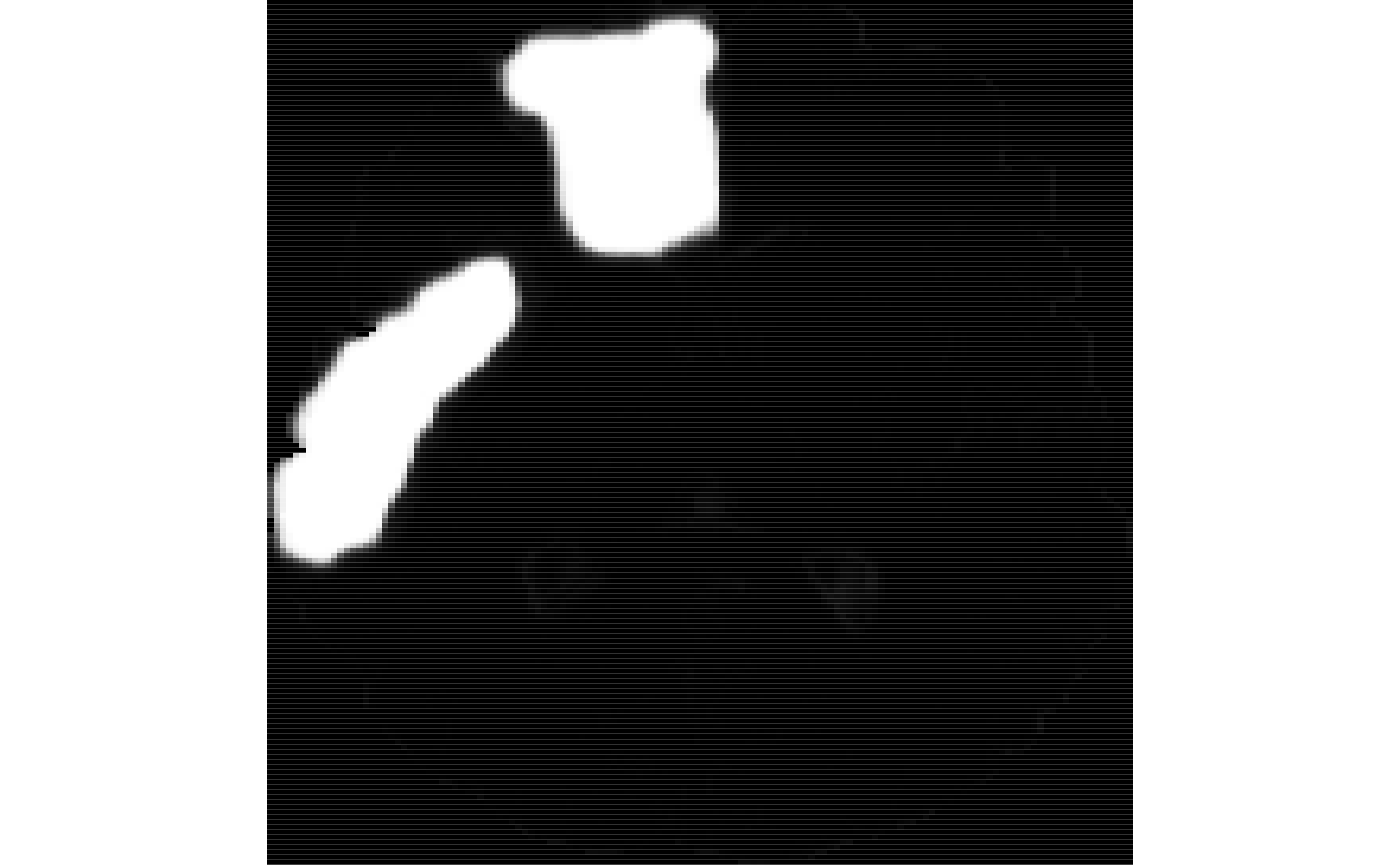
ti<-antsImageRead( getANTsRData("r27") ) timask=getMask(ti) seg2<-kmeansSegmentation( ti, 3 )$segmentation ll2<-simLesion( ti, 10, 6, myseed=919 ) # different sized lesion seg2[ ll2$lesion > 0.5 & seg2 > 0.5 ]<-4
Make training data
Create training data and map to the test subject. Note that a “real” application of this type would use cost function masking.
But let’s ignore that aspect of the problem here.
Make training data
img<-antsImageRead( getANTsRData("r16") ) seg<-kmeansSegmentation( img, 3 )$segmentation ll<-simLesion( img, 12, 5, myseed=1 ) seg[ ll$lesion > 0.5 & seg > 0.5 ]<-4
Pseudo-ground truth
This gives us a subject with a “ground truth” segmentation.
Now we get a new subject and map to the space of the arbitrarily chosen reference space.
The study
Perform training step
Now use these to train a model.
rad<-c(1,1) # fast setting mr<-c(1,2,4,2,1) # multi-res schedule, U-style schedule masks=list( getMask(seg), getMask(seg1) ) rfm<-mrvnrfs( list(seg,seg1) , list(list(ll$limg), list(ll1$limg) ), masks, rad=rad, nsamples = 500, ntrees=1000, multiResSchedule=mr, voxchunk=500, do.trace = 100)
## randomForest 4.6-14## Type rfNews() to see new features/changes/bug fixes.## ntree OOB 1 2 3 4
## 100: 8.60% 20.00% 7.49% 9.79% 3.59%
## 200: 8.50% 20.00% 7.49% 9.79% 3.19%
## 300: 9.20% 24.76% 7.82% 10.09% 3.19%
## 400: 9.10% 22.86% 7.82% 10.09% 3.59%
## 500: 9.10% 22.86% 7.82% 10.09% 3.59%
## 600: 9.20% 23.81% 7.82% 10.39% 3.19%
## 700: 9.10% 21.90% 8.14% 10.39% 3.19%
## 800: 9.10% 20.95% 8.47% 10.39% 3.19%
## 900: 9.00% 20.95% 8.14% 10.09% 3.59%
## 1000: 9.00% 20.95% 7.82% 10.39% 3.59%
## ntree OOB 1 2 3 4
## 100: 11.90% 18.56% 10.27% 16.22% 5.15%
## 200: 12.40% 18.56% 10.88% 16.52% 6.01%
## 300: 11.90% 17.53% 9.67% 16.52% 6.01%
## 400: 11.60% 16.49% 9.37% 16.52% 5.58%
## 500: 11.50% 16.49% 9.67% 15.93% 5.58%
## 600: 11.60% 16.49% 9.97% 15.63% 6.01%
## 700: 11.60% 17.53% 9.67% 15.63% 6.01%
## 800: 11.80% 18.56% 10.27% 15.34% 6.01%
## 900: 11.80% 16.49% 10.57% 15.63% 6.01%
## 1000: 11.90% 16.49% 9.97% 16.22% 6.44%
## ntree OOB 1 2 3 4
## 100: 6.90% 7.34% 8.25% 9.25% 1.66%
## 200: 6.40% 8.26% 7.30% 8.96% 0.83%
## 300: 6.70% 8.26% 7.62% 9.55% 0.83%
## 400: 6.60% 8.26% 7.30% 9.55% 0.83%
## 500: 6.70% 8.26% 7.62% 9.55% 0.83%
## 600: 6.80% 8.26% 7.30% 10.15% 0.83%
## 700: 6.80% 8.26% 7.30% 10.15% 0.83%
## 800: 6.80% 8.26% 7.30% 10.15% 0.83%
## 900: 6.80% 8.26% 7.30% 10.15% 0.83%
## 1000: 6.80% 8.26% 7.30% 10.15% 0.83%
## ntree OOB 1 2 3 4
## 100: 7.40% 12.38% 9.12% 8.93% 0.83%
## 200: 6.90% 10.48% 8.81% 8.04% 1.24%
## 300: 7.30% 12.38% 9.12% 8.33% 1.24%
## 400: 7.20% 13.33% 9.43% 7.44% 1.24%
## 500: 7.10% 13.33% 9.12% 7.44% 1.24%
## 600: 7.30% 13.33% 9.75% 7.44% 1.24%
## 700: 7.10% 13.33% 9.43% 7.14% 1.24%
## 800: 7.10% 13.33% 9.12% 7.44% 1.24%
## 900: 7.10% 13.33% 9.12% 7.44% 1.24%
## 1000: 7.20% 13.33% 9.12% 7.74% 1.24%
## ntree OOB 1 2 3 4
## 100: 6.80% 10.38% 8.62% 8.72% 0.40%
## 200: 7.00% 9.43% 8.92% 9.35% 0.40%
## 300: 6.90% 9.43% 8.92% 9.03% 0.40%
## 400: 6.80% 8.49% 9.54% 8.41% 0.40%
## 500: 7.10% 8.49% 9.85% 9.03% 0.40%
## 600: 6.90% 8.49% 9.54% 8.72% 0.40%
## 700: 6.90% 9.43% 9.23% 8.72% 0.40%
## 800: 7.00% 9.43% 9.54% 8.72% 0.40%
## 900: 7.00% 9.43% 9.54% 8.72% 0.40%
## 1000: 7.00% 9.43% 9.54% 8.72% 0.40%Combine with additional training runs
newrflist<-list() temp<-mrvnrfs( list(seg,seg1) , list(list(ll$limg), list(ll1$limg) ), masks, rad=rad, nsamples = 500, ntrees=1000, multiResSchedule=mr, voxchunk=500 ) for ( k in 1:length( mr ) ) if ( length( rfm$rflist[[k]]$classes ) == length( temp$rflist[[k]]$classes ) ) newrflist[[k]]<-combine( rfm$rflist[[k]], temp$rflist[[k]] ) rfm$rflist<-newrflist
Apply the model to new data
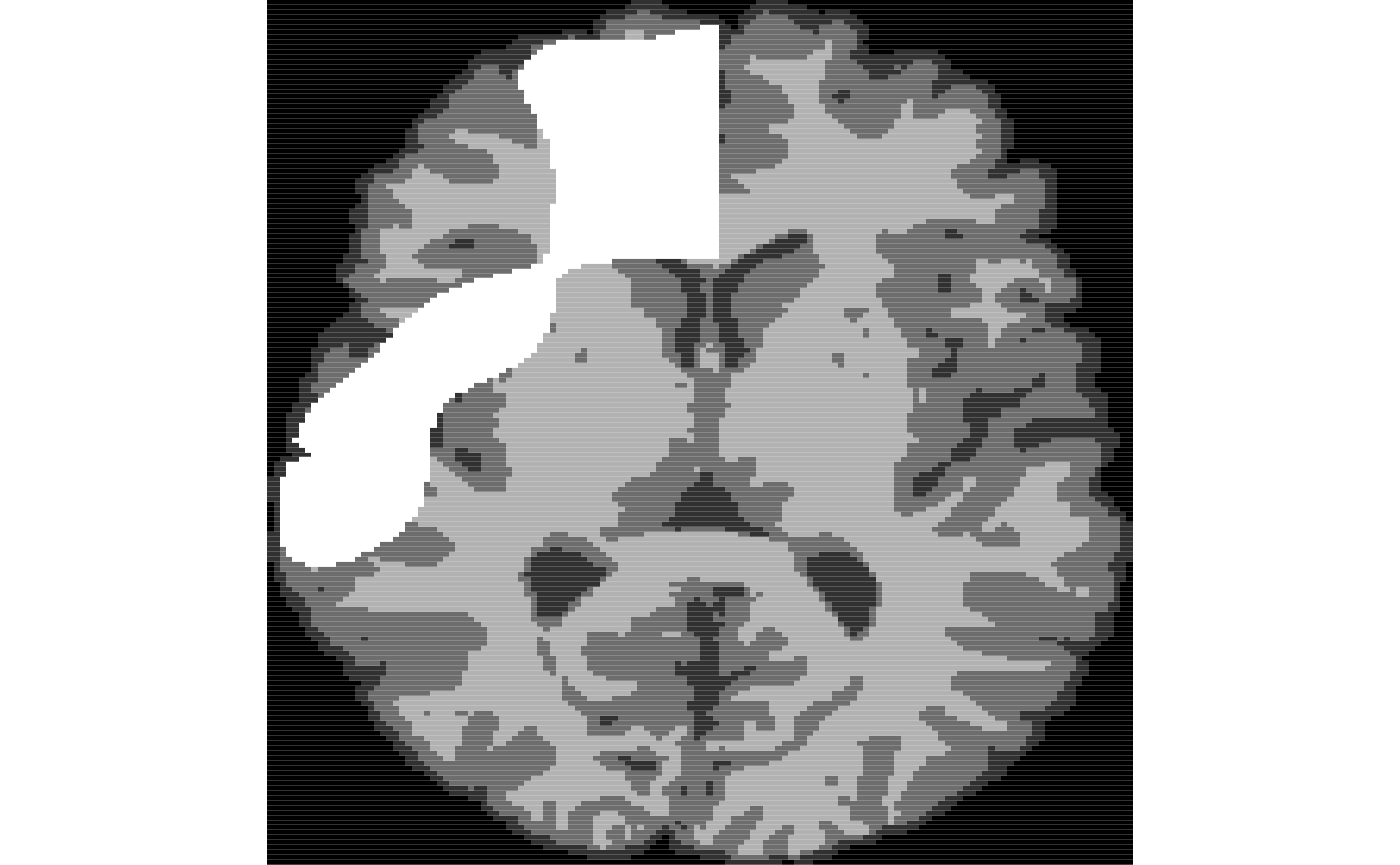
We apply the learned model to segment the new data.
mmseg<-mrvnrfs.predict( rfm$rflist, list(list(ll2$limg)), timask, rad=rad, multiResSchedule=mr, voxchunk=500 )